ARTICLE AD
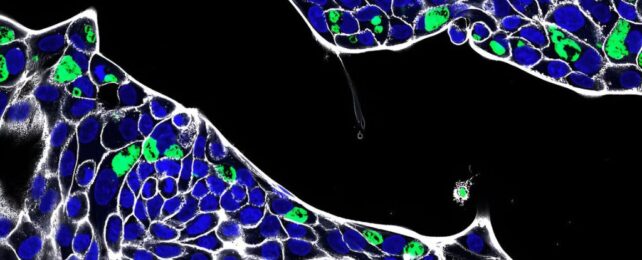 Stained human gastric cells infected with chlamydia (stained green). (Pargev Hovhannisyan/Universität Würzburg)
Stained human gastric cells infected with chlamydia (stained green). (Pargev Hovhannisyan/Universität Würzburg)
Chlamydia may have a secret hiding spot in the human body where it can avoid antibiotics.
Researchers at the University of Würzburg in Germany have found evidence that the human gut is an overlooked "natural niche" for this all too common sexually transmitted infection (STI).
If the team is right, it could help explain why Chlamydia trachomatis reinfections in treated patients often involve the exact same bacterial strain.
"It is therefore reasonable to assume that the bacteria find a niche in the body where they are not yet vulnerable, that they form a permanent reservoir there and can become active again later," explains chlamydia expert and microbiologist Thomas Rudel from the University of Würzburg.
That's a dangerous prospect, because the longer chlamydia sticks around in the body, the higher the risk of infertility issues and ovarian cancer. Oftentimes, a chlamydia infection causes no symptoms or only mild ones, such as pain, discharge, or itchiness. As a result, doctors recommend retesting for the STI three months after finishing a dose of antibiotics.
Chlamydia is the most common STI worldwide, so it is vital that scientists stay on top of effective treatments. If there really is a reservoir of bacteria hiding out in the body, it could result in growing antibiotic resistance.
To investigate further, Rudel and his colleagues in Germany studied three organoids, or mini cellular models: one of the stomach, one of the small intestine, and one of the colon or large intestine.
Though these self-organized clumps of human cells lacked a natural microbiota or an immune system, they were equipped with a blood supply and could demonstrate how chlamydia might infect gastrointestinal cells.
Ultimately, the team found that the tissue lining the inside of the human intestines was effective at keeping C. trachomatis out. Only when the tightly arranged cellular structure was damaged could the bacteria infiltrate.
If the bacteria made its way into blood circulation, however, researchers found chlamydia repeatedly bypassed the gut's physical barrier and established an infection in deeper parts.
This suggests that chlamydia can infect the gut very easily via the blood but not so easily via the intestinal lining. Clinical trials are now needed to confirm that idea.
"This is the first report of C. trachomatis infection in human primary intestinal epithelial cells," write lead author microbiologist Pargev Hovhannisyan and his colleagues, "supporting a possible niche for chlamydial infection in the human intestinal tissue."
Chlamydia is known to infect a wide range of cell types, including those in the cervix, urethra, testicles, mouth, throat, anus, and eyes. But most experiments on the bacteria so far have focused on its infection in the genital tract.
Some studies, however, have found the DNA of C. trachomitis in the intestinal biopsies of patients, and this suggests the infection can penetrate deeper into the body than we realized.
While C. trachomitis is a human-specific pathogen, other species of chlamydia that can infect wild or domesticated animals can, and often do, make it to the gastrointestinal tract.
In fact, when mice are orally inoculated with the human-specific pathogen, chlamydia cells cross the gastrointestinal barrier and establish "long lasting non-pathological colonization in the large intestine."
Non-pathological means though the bacteria are alive inside the cells they aren't actively replicating. When threatened by antibiotics or other stressful events, the microbes can enter a dormant state to survive, reawakening only when conditions are appropriate to seed new infections elsewhere.
As a result, some scientists have proposed that the human GI tract is a possible secret hiding spot for persistent chlamydial infections, and the new experiments support that hypothesis.
The team now wants to test which cells in the gut are most susceptible to chlamydia infection, and why.
The study was published in PLOS Pathogens.

 2 months ago
21
2 months ago
21